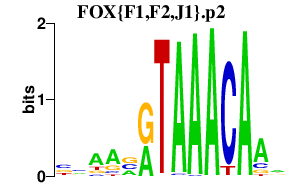
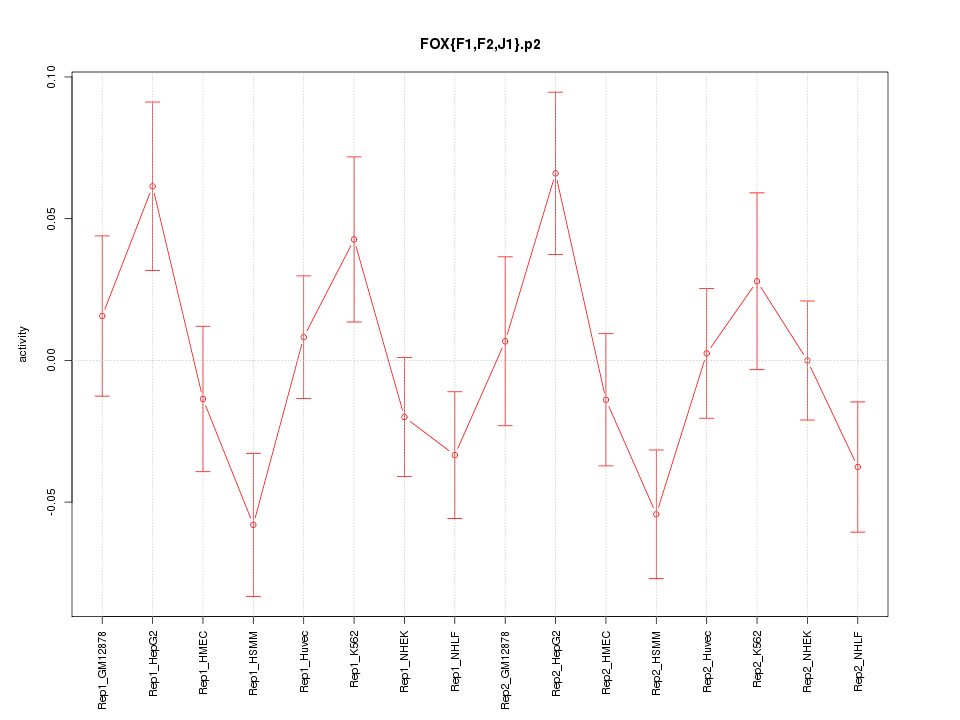
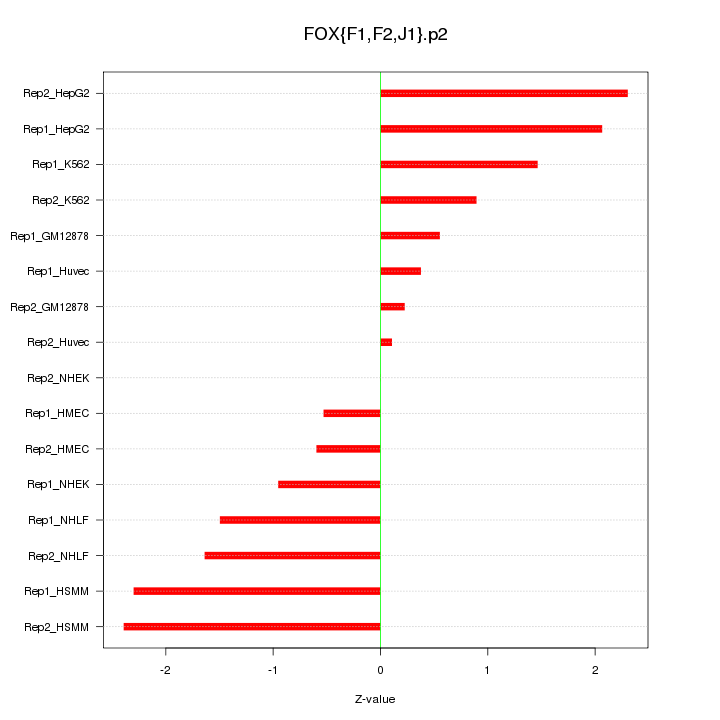
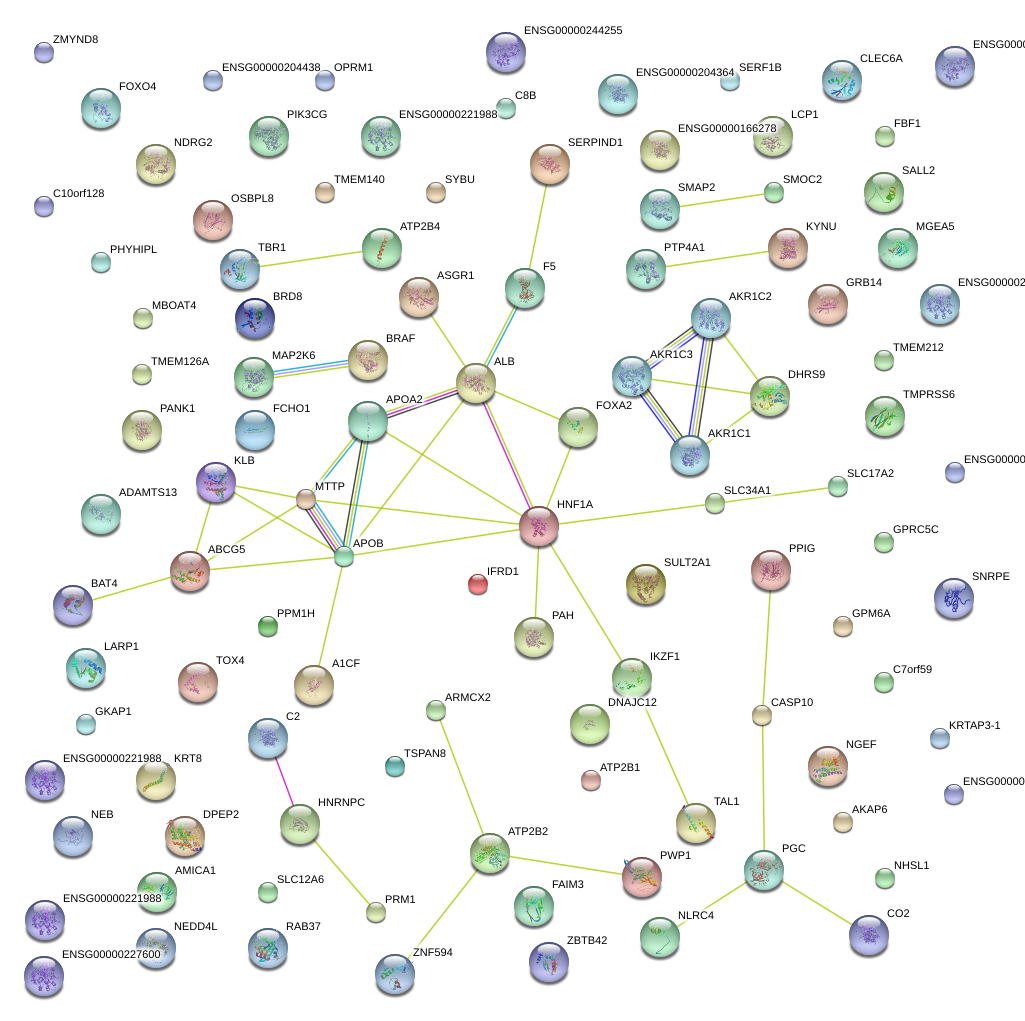
|
chr1_-_159459814
|
3.846
|
|
APOA2
|
apolipoprotein A-II
|
|
chr1_-_159459999
|
3.763
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr12_-_51629874
|
3.193
|
|
KRT8
|
keratin 8
|
|
chr17_+_69939130
|
3.121
|
NM_022036
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr14_-_20562962
|
2.884
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr1_-_47468029
|
2.742
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr22_+_19458378
|
2.642
|
NM_000185
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chr5_-_42847716
|
2.397
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr17_-_7023283
|
2.384
|
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr7_+_134483305
|
2.230
|
NM_018295
|
TMEM140
|
transmembrane protein 140
|
|
chr20_-_45410033
|
2.129
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr13_+_49658016
|
2.086
|
|
|
|
|
chr11_-_117589283
|
1.932
|
NM_153206
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1
|
|
chr2_-_233586145
|
1.848
|
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr2_-_233586164
|
1.736
|
NM_019850
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr12_+_119900931
|
1.727
|
NM_000545
|
HNF1A
|
HNF1 homeobox A
|
|
chr17_-_7023378
|
1.673
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr12_+_119900677
|
1.668
|
|
HNF1A
|
HNF1 homeobox A
|
|
chr12_-_101835026
|
1.665
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr6_-_41823056
|
1.582
|
NM_001166424
NM_002630
|
PGC
|
progastricsin (pepsinogen C)
|
|
chr13_-_45654292
|
1.581
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr4_+_100714995
|
1.572
|
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr1_-_167822248
|
1.571
|
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|
|
chr15_-_32398220
|
1.570
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr1_+_116455898
|
1.536
|
NM_152367
|
MAB21L3
|
mab-21-like 3 (C. elegans)
|
|
chr16_-_11282595
|
1.527
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr13_-_45654327
|
1.477
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr14_+_104337923
|
1.467
|
NM_001137601
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr13_-_45654296
|
1.464
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr19_-_53081326
|
1.445
|
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chr10_-_91395194
|
1.432
|
NM_148977
|
PANK1
|
pantothenate kinase 1
|
|
chr14_+_31868229
|
1.405
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr1_-_1346465
|
1.403
|
NM_001145210
|
LOC441869
|
hypothetical protein LOC441869
|
|
chr2_-_43919392
|
1.388
|
NM_022436
|
ABCG5
|
ATP-binding cassette, sub-family G (WHITE), member 5
|
|
chr6_-_138862271
|
1.384
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr14_+_104338423
|
1.356
|
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr10_-_69267774
|
1.351
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr8_-_30121741
|
1.343
|
NM_001100916
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4
|
|
chr6_-_138862273
|
1.334
|
|
|
|
|
chr1_-_167822334
|
1.320
|
NM_000130
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|
|
chr7_+_106292975
|
1.275
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr2_-_165133239
|
1.263
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr7_+_50318801
|
1.248
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr3_+_173043832
|
1.242
|
NM_001164436
|
TMEM212
|
transmembrane protein 212
|
|
chr10_-_52315390
|
1.203
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr22_-_35835548
|
1.198
|
|
TMPRSS6
|
transmembrane protease, serine 6
|
|
chr16_-_66590856
|
1.195
|
NM_022355
|
DPEP2
|
dipeptidase 2
|
|
chr6_+_32003180
|
1.190
|
NM_000063
NM_001145903
|
C2
|
complement component 2
|
|
chr1_+_40635058
|
1.173
|
NM_001198980
|
SMAP2
|
small ArfGAP2
|
|
chr2_+_143351695
|
1.165
|
|
KYNU
|
kynureninase
|
|
chr18_+_53863865
|
1.158
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr2_-_32344192
|
1.157
|
NM_021209
|
NLRC4
|
NLR family, CARD domain containing 4
|
|
chr2_+_169637255
|
1.154
|
NM_001142270
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr19_+_17723285
|
1.137
|
NM_001161359
NM_015122
|
FCHO1
|
FCH domain only 1
|
|
chr12_-_61615124
|
1.135
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr4_+_74488910
|
1.123
|
|
ALB
|
albumin
|
|
chr6_-_31740466
|
1.111
|
|
GPANK1
|
G patch domain and ankyrin repeats 1
|
|
chr10_-_5035967
|
1.078
|
NM_001135241
|
AKR1C1
AKR1C2
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr4_+_74488835
|
1.077
|
NM_000477
|
ALB
|
albumin
|
|
chr8_-_110724594
|
1.076
|
NM_001099756
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr4_-_176971175
|
1.076
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr12_-_69837831
|
1.074
|
|
TSPAN8
|
tetraspanin 8
|
|
chr3_-_10522267
|
1.070
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr20_-_22514092
|
1.064
|
NM_153675
|
FOXA2
|
forkhead box A2
|
|
chr6_+_64289585
|
1.063
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr1_-_57204208
|
1.045
|
NM_000066
|
C8B
|
complement component 8, beta polypeptide
|
|
chr17_+_64922420
|
1.038
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr10_+_60606352
|
1.038
|
NM_032439
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like
|
|
chr9_-_85622366
|
1.037
|
NM_001135953
NM_025211
|
GKAP1
|
G kinase anchoring protein 1
|
|
chr7_+_106293112
|
1.028
|
NM_002649
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr5_+_154072654
|
1.021
|
NM_015315
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr10_-_52315436
|
1.015
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr7_-_140270749
|
1.010
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr6_-_26038817
|
0.984
|
NM_005835
|
SLC17A2
|
solute carrier family 17 (sodium phosphate), member 2
|
|
chr2_+_161980865
|
0.965
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr7_+_111850367
|
0.962
|
NM_001007245
NM_001197080
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr2_+_201756048
|
0.944
|
|
CASP10
|
caspase 10, apoptosis-related cysteine peptidase
|
|
chr2_-_21084077
|
0.941
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr1_-_205161821
|
0.926
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr17_+_70244945
|
0.925
|
NM_001006638
NM_001163990
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chrX_+_70232362
|
0.924
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr2_+_143351536
|
0.922
|
NM_001032998
NM_003937
|
KYNU
|
kynureninase
|
|
chr4_+_39084867
|
0.919
|
NM_175737
|
KLB
|
klotho beta
|
|
chr3_-_10522340
|
0.900
|
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr6_+_154373323
|
0.896
|
NM_001145279
NM_001145280
NM_001145281
|
OPRM1
|
opioid receptor, mu 1
|
|
chr17_-_36418869
|
0.892
|
NM_031958
|
KRTAP3-1
|
keratin associated protein 3-1
|
|
chr10_+_4995597
|
0.890
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr9_+_135276940
|
0.887
|
NM_139025
NM_139026
NM_139027
|
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13
|
|
chr10_+_4995453
|
0.884
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chrX_-_153698930
|
0.884
|
|
HMGN1P37
|
high-mobility group nucleosome binding domain 1 pseudogene 37
|
|
chr10_-_50066386
|
0.883
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr6_+_32230172
|
0.883
|
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr7_-_25234582
|
0.876
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr4_+_69716298
|
0.875
|
NM_001075
NM_001144767
|
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr2_-_87156616
|
0.873
|
|
LOC285074
|
anaphase promoting complex subunit 1 pseudogene
|
|
chr17_-_23721485
|
0.869
|
NM_000638
|
VTN
|
vitronectin
|
|
chr6_+_32229599
|
0.868
|
NM_005155
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 read-through transcript
palmitoyl-protein thioesterase 2
|
|
chr4_+_22608252
|
0.860
|
|
|
|
|
chr12_-_94914133
|
0.857
|
NM_002108
|
HAL
|
histidine ammonia-lyase
|
|
chr18_-_50777634
|
0.852
|
NM_025214
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr1_+_196874867
|
0.852
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr17_-_23721468
|
0.841
|
|
VTN
|
vitronectin
|
|
chr22_+_29696662
|
0.841
|
|
|
|
|
chr2_-_135498716
|
0.831
|
NM_001018046
NM_025052
|
YSK4
|
YSK4 Sps1/Ste20-related kinase homolog (S. cerevisiae)
|
|
chr6_+_44352443
|
0.826
|
|
TMEM151B
|
transmembrane protein 151B
|
|
chr2_+_86800940
|
0.824
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr13_-_48873735
|
0.818
|
NM_030925
|
CAB39L
|
calcium binding protein 39-like
|
|
chr10_+_102722275
|
0.817
|
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr4_+_88939725
|
0.815
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr4_+_39084936
|
0.811
|
|
KLB
|
klotho beta
|
|
chr19_+_18357966
|
0.807
|
NM_004864
|
GDF15
|
growth differentiation factor 15
|
|
chr2_+_201755865
|
0.802
|
NM_001230
NM_032974
NM_032977
|
CASP10
|
caspase 10, apoptosis-related cysteine peptidase
|
|
chr2_+_176761552
|
0.801
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr2_+_43919606
|
0.797
|
NM_022437
|
ABCG8
|
ATP-binding cassette, sub-family G (WHITE), member 8
|
|
chr4_-_123761615
|
0.795
|
NM_021803
|
IL21
|
interleukin 21
|
|
chr3_-_71376919
|
0.793
|
|
FOXP1
|
forkhead box P1
|
|
chr14_+_94148466
|
0.793
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr12_-_69838045
|
0.792
|
NM_004616
|
TSPAN8
|
tetraspanin 8
|
|
chr5_-_169657360
|
0.791
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr15_-_38387410
|
0.790
|
NM_004573
|
PLCB2
|
phospholipase C, beta 2
|
|
chr11_-_114880275
|
0.781
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr12_+_130004404
|
0.781
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr5_+_137701122
|
0.780
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr8_+_82807218
|
0.779
|
NM_152284
|
CHMP4C
|
chromatin modifying protein 4C
|
|
chr10_+_4995685
|
0.773
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr19_-_51979838
|
0.768
|
NM_001145145
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5
|
|
chr6_+_30238961
|
0.768
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr6_+_112481970
|
0.766
|
NM_003880
NM_198239
|
WISP3
|
WNT1 inducible signaling pathway protein 3
|
|
chr4_-_186969041
|
0.762
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chrX_-_48578878
|
0.762
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr5_-_169657395
|
0.757
|
NM_005565
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr20_-_42561725
|
0.756
|
|
|
|
|
chr19_+_15612706
|
0.750
|
NM_000896
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3
|
|
chr4_+_172971149
|
0.740
|
NM_001034845
|
GALNTL6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6
|
|
chr5_+_171553780
|
0.737
|
NM_001171183
|
EFCAB9
|
EF-hand calcium binding domain 9
|
|
chr5_-_169657312
|
0.737
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr9_+_135277264
|
0.737
|
|
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13
|
|
chr6_-_50025115
|
0.733
|
NM_001166478
|
DEFB133
|
defensin, beta 133
|
|
chr14_-_22358752
|
0.728
|
NM_001126106
|
SLC7A7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7
|
|
chr15_-_38387357
|
0.728
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr1_+_28134090
|
0.726
|
NM_001009568
NM_014474
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B
|
|
chr9_-_115880494
|
0.723
|
NM_001633
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr4_+_35962038
|
0.720
|
NM_001170700
|
DTHD1
|
death domain containing 1
|
|
chr17_-_43390108
|
0.719
|
NM_024320
|
PRR15L
|
proline rich 15-like
|
|
chr1_+_190811479
|
0.718
|
NM_002922
|
RGS1
|
regulator of G-protein signaling 1
|
|
chr2_-_180319013
|
0.716
|
NM_001113397
|
ZNF385B
|
zinc finger protein 385B
|
|
chr6_-_25834733
|
0.715
|
NM_170745
|
HIST1H2AA
|
histone cluster 1, H2aa
|
|
chr17_-_23721430
|
0.713
|
|
VTN
|
vitronectin
|
|
chr5_-_59819642
|
0.712
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_169657192
|
0.709
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr1_+_144149794
|
0.708
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr1_-_24067347
|
0.701
|
NM_000147
|
FUCA1
|
fucosidase, alpha-L- 1, tissue
|
|
chr16_+_11346780
|
0.697
|
NM_152308
|
C16orf75
|
chromosome 16 open reading frame 75
|
|
chr9_-_205892
|
0.696
|
NM_152569
|
C9orf66
|
chromosome 9 open reading frame 66
|
|
chr18_+_30327341
|
0.694
|
|
DTNA
|
dystrobrevin, alpha
|
|
chr4_+_187424336
|
0.690
|
|
F11
|
coagulation factor XI
|
|
chrX_+_57329834
|
0.687
|
NM_174912
|
FAAH2
|
fatty acid amide hydrolase 2
|
|
chr17_-_23721420
|
0.685
|
|
VTN
|
vitronectin
|
|
chr7_+_149733309
|
0.685
|
|
|
|
|
chr9_-_115880366
|
0.682
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr7_+_77307382
|
0.667
|
NM_001127357
NM_001127359
NM_020432
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr17_+_38178969
|
0.662
|
NM_032353
|
VPS25
|
vacuolar protein sorting 25 homolog (S. cerevisiae)
|
|
chr17_-_23721351
|
0.661
|
|
VTN
|
vitronectin
|
|
chr1_+_198278339
|
0.658
|
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr2_-_39201631
|
0.654
|
|
SOS1
|
son of sevenless homolog 1 (Drosophila)
|
|
chr17_+_69950782
|
0.654
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr15_-_38387164
|
0.653
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr11_+_22644732
|
0.645
|
NM_001143830
|
GAS2
|
growth arrest-specific 2
|
|
chr14_-_20636568
|
0.645
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chr20_+_23293020
|
0.642
|
NM_022482
|
GZF1
|
GDNF-inducible zinc finger protein 1
|
|
chr9_-_115880281
|
0.638
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr12_-_31370305
|
0.638
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr18_+_53170718
|
0.633
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr5_+_147754467
|
0.631
|
|
FBXO38
|
F-box protein 38
|
|
chr2_+_86800898
|
0.631
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr2_+_86801081
|
0.628
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr16_+_716936
|
0.616
|
NM_207112
NM_032304
|
HAGHL
|
hydroxyacylglutathione hydrolase-like
|
|
chr1_-_24342158
|
0.615
|
NM_021258
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr10_-_54201400
|
0.615
|
NM_000242
|
MBL2
|
mannose-binding lectin (protein C) 2, soluble
|
|
chr11_-_116213511
|
0.605
|
NM_000039
|
APOA1
|
apolipoprotein A-I
|
|
chr17_-_73636386
|
0.604
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr4_+_187424111
|
0.598
|
NM_000128
|
F11
|
coagulation factor XI
|
|
chr17_+_75366524
|
0.597
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr1_-_31117948
|
0.593
|
|
SDC3
|
syndecan 3
|
|
chr6_+_32229771
|
0.590
|
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 read-through transcript
palmitoyl-protein thioesterase 2
|
|
chr11_-_102331643
|
0.584
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chr5_-_110090264
|
0.583
|
NM_001039763
|
TMEM232
|
transmembrane protein 232
|
|
chr11_-_16953135
|
0.582
|
|
RPL36A
|
ribosomal protein L36a
|
|
chr11_-_63690069
|
0.577
|
NM_014067
|
MACROD1
|
MACRO domain containing 1
|
|
chr18_+_30327243
|
0.576
|
NM_001198945
NM_001392
NM_032975
NM_032979
|
DTNA
|
dystrobrevin, alpha
|
|
chr4_-_38710376
|
0.570
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr16_+_11346812
|
0.569
|
|
C16orf75
|
chromosome 16 open reading frame 75
|
|
chr1_+_74473672
|
0.566
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr14_-_99140195
|
0.566
|
NM_001144995
|
CCDC85C
|
coiled-coil domain containing 85C
|
|
chr7_+_106292932
|
0.560
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr17_-_72151314
|
0.559
|
NM_018414
|
ST6GALNAC1
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1
|
|
chr10_+_70517863
|
0.558
|
|
SRGN
|
serglycin
|
|
chr12_-_31370351
|
0.555
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr12_+_77963563
|
0.552
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr10_+_70517823
|
0.552
|
NM_002727
|
SRGN
|
serglycin
|